Large Ribozymes
Ribozymes can be classified based on their size and functional complexity. Large ribozymes, which range from several hundred to thousands of nucleotides, include Ribonuclease P (RNase P), group I and II introns, and the catalytic components of complex ribonucleoprotein machines such as the spliceosome and ribosome. While these ribozymes typically function in conjunction with proteins in vivo, some, like RNase P and group I and II introns, are capable of catalysis on their own in vitro, although often with reduced efficiency.
Key characteristics of large ribozymes:
- Metal ion dependence: Catalytic activity typically requires divalent metal ions, primarily Mg²⁺, which stabilize the ribozyme structure and participate in catalysis.
- Reaction mechanism: These ribozymes catalyze reactions that generate products with 3' hydroxyl and 5' phosphate groups, essential for RNA processing.
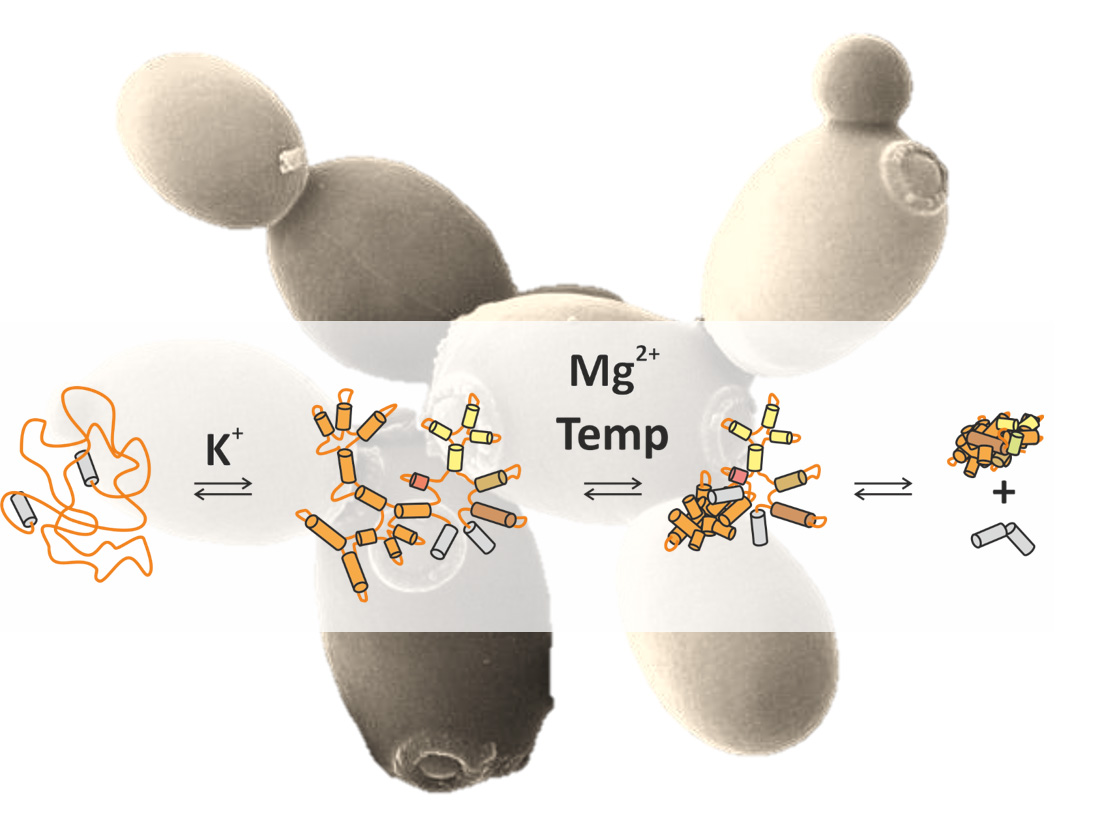
- Complex structural organization: Extensive tertiary interactions give rise to highly structured domain architectures, essential for their function.
Group II Introns: evolutionary ancestors of the spliceosome
Group II introns, which range in length from 400 to 1000 nucleotides, are thought to be the evolutionary ancestors of the spliceosome, the cellular machinery responsible for pre-mRNA splicing. These self-splicing ribozymes excise themselves from precursor mRNAs, leaving behind properly processed transcripts. In addition to their biological significance, group II introns have great potential for applications in biotechnology and gene therapy, such as site-specific genome engineering. A well-characterized example is the Sc.ai5γ intron from Saccharomyces cerevisiae, which adopts a defined three-dimensional structure and efficiently catalyzes its own excision. Our research aims to understand the intricate mechanisms of both splicing and folding in these ribozymes.
To achieve this, we employ:
- Fluorescence-based approaches such as single-molecule Förster resonance energy transfer (smFRET) to track RNA folding in real time and assess catalytic activity.
- Structural techniques including NMR spectroscopy, X-ray crystallography, and molecular dynamics (MD) simulations to gain deeper insights into key structural elements governing function.
By combining biochemical, biophysical, and computational methods, we aim to elucidate the fundamental principles that govern ribozyme activity, ultimately contributing to a better understanding of RNA catalysis and its evolutionary implications.
Literature
Fabio D. Steffen, Mokrane Khier, Danny Kowerko, Richard A. Cunha, Richard Börner, and Roland K.O. Sigel, Nat. Commun, 2020, 11, 104.
doi:10.1038/s41467-019-13683-4
Bishnu P. Paudel, Erica Fiorini, Richard Börner, Roland K.O. Sigel, and David S. Rueda, Proc. Natl. Acad. Sci. USA, 2018, 115, 11917-11922.
doi:10.1073/pnas.1806685115
Miriam Skilandat, Magdalena Rowinska-Zyrek, Roland K. O. Sigel*, RNA, 2016, 22, 750-763.
doi:10.1261/rna.053843.115
Erica Fiorini, Richard Börner, and Roland K.O. Sigel., CHIMIA, 2015, 69, 207-212.
doi:10.2533/chimia.2015.207
Anita G. Schmitz, Susann Zelger‐Paulus, Gilles Gasser, and Roland K.O. Sigel, ChemBioChem, 2015, 16, 1302-1306.
doi:10.1002/cbic.201500180
Miriam Steiner, David Rueda, Roland K.O. Sigel, Angew. Chem. Int. Ed. Engl., 2009, 48, 13853-13858.
doi:10.1002/anie.200903809
doi:10.1073/pnas.0809940105
Michèle C. Erat, Roland K. O. Sigel, J. Biol. Inorg. Chem., 2008, 13, 1025-1036.
doi:10.1007/s00775-008-0390-7